Article Text
Abstract
Background In the face of the COVID-19 pandemic, the Defence Science and Technology Laboratory (Dstl) and Defence Pathology combined to form the Defence Clinical Lab (DCL), an accredited (ISO/IEC 17025:2017) high-throughput SARS-CoV-2 PCR screening capability for military personnel.
Laboratory structure and resource The DCL was modular in organisation, with laboratory modules and supporting functions combining to provide the accredited SARS-CoV-2 (envelope (E)-gene) PCR assay. The DCL was resourced by Dstl scientists and military clinicians and biomedical scientists.
Laboratory results Over 12 months of operation, the DCL was open on 289 days and tested over 72 000 samples. Six hundred military SARS-CoV-2-positive results were reported with a median E-gene quantitation cycle (Cq) value of 30.44. The lowest Cq value for a positive result observed was 11.20. Only 64 samples (0.09%) were voided due to assay inhibition after processing started.
Conclusions Through a sustained effort and despite various operational issues, the collaboration between Dstl scientific expertise and Defence Pathology clinical expertise provided the UK military with an accredited high-throughput SARS-CoV-2 PCR test capability at the height of the COVID-19 pandemic. The DCL helped facilitate military training and operational deployments contributing to the maintenance of UK military capability. In offering a bespoke capability, including features such as testing samples in unit batches and oversight by military consultant microbiologists, the DCL provided additional benefits to the UK Ministry of Defence that were potentially not available from other SARS-CoV-2 PCR laboratories. The links between Dstl and Defence Pathology have also been strengthened, benefitting future research activities and operational responses.
- COVID-19
- diagnostic microbiology
- molecular diagnostics
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
WHAT IS ALREADY KNOWN ON THIS TOPIC
The ability of Defence Science and Technology Laboratory and Defence Pathology to combine and rapidly develop an accredited, high-throughput, clinical diagnostic PCR assay was untested before the capability was successfully developed and operated.
WHAT THIS STUDY ADDS
SARS-CoV-2 (envelope gene) PCR quantitation cycle values indicated wide differences in viral loads in the largely asymptomatic individuals tested.
Factors such as the lack of a centralised sample booking system accessible to the laboratory, units submitting samples, and a UK Ministry of Defence co-ordination cell increased laboratory burdens.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Any similar future capability could be improved by considering this and other factors such as uniform packaging and sampling consumables.
Introduction
SARS-CoV-2 is an RNA genome virus belonging to the Coronaviridae and is the causative agent of the COVID-19 pandemic.1 2 SARS-CoV-2 is classified in the UK as an Advisory Committee on Dangerous Pathogens Hazard Group 3 pathogen; however, guidance3 4 permits non-propagative diagnostic testing to be carried out at containment level 2 (CL2) with non-inactivated samples being handled within microbiological safety cabinets (MSCs). Real-time reverse transcriptase PCR from oro or naso pharyngeal (OP/NP) swab samples is typically used to detect SARS-CoV-2 in high-throughput laboratories.
In March 2020, Defence Science and Technology Laboratory (Dstl) repurposed CL2 research laboratories to provide a high-throughput SARS-CoV-2 PCR capability to help meet testing requirements in the face of the pandemic. Staff were assembled with existing expertise plus experience from deployment to NHS labs early in the pandemic or to Sierra Leone during the 2013–2016 Ebola outbreak.5 Previous Dstl research projects also helped facilitate the development of the capability.
In April 2020, with available testing capacity at Dstl and a requirement to maintain military operations during the pandemic, Dstl formed a partnership with Defence Pathology to form the Defence Clinical Lab (DCL). This partnership provided clinical governance, military experience and laboratory assistance from military biomedical scientists (BMSs).
From May to August 2020, the DCL tested sporadic military samples. From September 2020, under contract with the Department of Health and Social Care, the DCL provided a high-throughput testing capability for military personnel and their families to facilitate operational deployments, overseas postings, training and work-related travel across all three UK services. Several Dstl and Ministry of Defence (MoD) research trials were also supported.
Taskings and guidance for military units
The DCL provided asymptomatic cohort screening and occasional symptomatic testing from NP swab samples (in viral transport medium (VTM) or universal transport medium (UTM)). Testing requests were directed via Permanent Joint Headquarters (PJHQ) COVID-19 Co-ordinating (Co-ord) Cell from taskings that arrived via a quarantine facility, test task order or defence primary healthcare unit. The PJHQ Co-ord Cell assessed and prioritised requests and then liaised with DCL operations managers to plan testing. Tasking requests were sent from units as a nominal roll, a Microsoft Excel spreadsheet which requested four personal identifiers be supplied per sample (surname, forename, date of birth and service number). Contact details of requesting medical officers (MOs), to facilitate reporting under Caldicott principles,6 were also requested. Guidance on how to package samples to the UN3373 standard was provided. Samples were transported to Dstl by military or commercial drivers. MoD Police helped facilitate a 24-hour reception facility at Dstl with samples delivered overnight for next day testing.
Laboratory structure and supporting functions
The DCL was modular (Figure 1) with individual leads assigned to develop and maintain each module and supporting function, in conjunction with a leadership team. Depending on sample numbers, 16–19 Dstl staff and military scientists resourced each shift, with staggered start and finish times. Typically, the DCL was open from 08:00 to 19:00–22:00 7 days a week, with a 13:30 sample delivery cut-off beyond which testing that day was not guaranteed. Occasional requirements to process above the agreed 500-sample per day capacity were incorporated by increasing staff numbers. Turnaround times for results varied, depending on the urgency of samples with those marked as D-1 (ie, deployment in 1 day) being prioritised.
Overview of DCL structure and operations. Top: supporting functions; bottom: laboratory process and reporting (overlain over arrow). DCA, Defence Consultant Advisor, DCL, Defence Clinical Lab; Dstl, Defence Science and Technology Laboratory; LIMS, laboratory information management system; MSC, microbiological safety cabinet; OpSec, operational security; PerSec, personal security.
Laboratory structure
Lysis buffer preparation
In a ‘clean’ laboratory (reducing contamination risks), RNA extraction lysis buffers were prepared each morning. MS2 bacteriophage, a non-pathogenic virus with RNA genome, was added to buffers at a precise concentration to incorporate a RNA extraction control for each sample. Two different RNA extraction platforms (see RNA extraction) were used with different lysis buffers prepared for each platform.
Admin and laboratory information management system (LIMS)
A bespoke LIMS was developed against DCL processes (taskings, sample reception, result upload, reporting and statistics). The database underlying the LIMS was built with Oracle Database V.12c. A browser-based application, used by laboratory staff to access the database, was developed with Oracle APEX. The LIMS user application included an import wizard to automatically load data from the nominal roll spreadsheet. If samples arrived at the DCL with an incorrectly formatted nominal roll, the application also permitted manual input of information. Four barcoded labels for each sample were printed and sent to sample reception. The display included visual alerts if the LIMS detected certain anomalies (eg, impossible date of birth). The LIMS user application also included a second import wizard for automatically loading data files generated by the autovalidation script (see the PCR section).
Sample reception
Samples were removed from tertiary packaging and checked against the nominal to ensure expected samples were present. Mismatches in patient information with that supplied on the nominal—when visible on tubes through the secondary packaging—were recorded as deviations and reported to admin, which tried to rectify them with the requestor. The swab tube (within secondary packaging) and barcoded labels were boxed for transfer to sample preparation. For the first 7 months of operations, sample reception was conducted on a laboratory bench but after several instances of samples arriving in incorrect packaging, a screening procedure was introduced where tertiary packaging was opened within a class 1 MSC to ensure they were packaged safely.
Sample preparation
Samples received from reception were unpacked and processed within a class 1 MSC. The cabinet requirement and operative burden meant that this module required the highest laboratory and staff resource. Tubes were unpacked and rechecked for leaks, vortexed and held (10 min). Patient information on each tube was matched with the barcoded label and deviations were recorded on a form held within the cabinet. This was photographed and electronically sent to admin, which rectified deviations with the requestor. Samples with unresolvable deviations were destroyed.
To process the sample, a lysis buffer tube and an empty tube were labelled with barcoded labels. The VTM/UTM sample was then transferred into the empty tube and 200 µL of this was subsequently pipetted into the lysis buffer tube. The remaining sample was held as a freezer aliquot (for repeat testing). Up to 42 samples were processed per MSC batch, with two negative extraction controls.
Sample lysis buffer tubes were submerged in 10 000 ppm sodium hypochlorite, removed from the MSC, dried and placed in a benchtop heat block which heated tubes to 68°C for 15 min. The SARS-CoV-2 inactivation efficacy of lysis buffers from kits used on both extraction platforms was verified,7 with the heat step ensuring total inactivation.
Samples were then scanned into specific positions on a 96-well rack and a plate plan saved and printed. Samples remained in well positions for the remaining laboratory process (RNA extraction and PCR). Eighty-four samples and four controls were processed on each plate. Freezer aliquot tubes were scanned into position in a box and stored at −80°C.
RNA extraction
A witnessed process verified that eight samples on each 96-well plate were in the correct position and, within an MSC, the inactivated sample mixes were transferred by multichannel pipette to the specific 96-well consumable used by the RNA extraction platform. To ensure resilience, in the face of global reagent shortages, two 96-well RNA extraction platforms, the Kingfisher Flex (ThermoFisher) and the QiaCube HT (Qiagen), were used. On the Qiacube HT, samples were processed using the QIAamp 96 Virus QIAcube HT Kit and on the Kingfisher Flex by the 5× Magmax Pathogen RNA/DNA kit. Run times between platforms varied: ~90 min for QiaCube HT and 32 min for Kingfisher Flex.
PCR
In early 2020, Public Health England (PHE) published a PCR protocol describing the use of the RNA-dependent RNA polymerase (RdRp) PCR assay for detection of SARS-CoV-2. The publication in which this PCR assay was first described8 also included description of an envelope (E) gene assay. In the first 2 weeks of DCL development both assays were evaluated as screening PCRs. Assays were multiplexed with a MS2 bacteriophage assay to form the RNA extraction control,9 and formulated, by an in-house Dstl PCR reagent team, in the Taqman Fastvirus 1-step mastermix (ThermoFisher) shown previously by Dstl to have inhibitor tolerance,.10 The E-gene assay demonstrated increased performance (in terms of analytical and diagnostic sensitivity and the ease with which it multiplexed with the MS2 PCR) over the RdRP assay. With similar findings reported elsewhere,11 the E-gene assay was selected. The multiplexed E-gene and MS2 assay was premade and stored into 96-well PCR plates.
RNA extracts, brought through in 96-well format, were transferred into position on these plates by multichannel pipettes in a witnessed process and run as a 45-cycle PCR on QuantStudio 7Flex PCR platforms (ThermoFisher).
Each plate contained RNA extracts from up to 84 samples and four naïve lysis buffers (acting as negative extraction controls). PCR positive and negative controls were added to remaining wells. Following a visual technical validation of results, an autovalidation script (written in R V.3.6.1, https://cran.r-project.org/) analysed results. This script screened data (from a manually exported .csv file generated by QuantStudio software) ensuring PCR quantitation cycle (Cq) values from RNA extraction and PCR control wells were within set boundaries. The script then analysed MS2 internal control Cq values from each sample. Samples outside a Cq range of 15–33 were flagged as inhibited and a repeat test was performed. Void samples (outside this range after retest) were reported as indeterminate and a further sample was requested.
Finally, the script analysed E-gene data. Results were reported as SARS-CoV-2 RNA not detected; SARS-CoV-2 RNA detected or indeterminate. All results with Cq above 42 were reported SARS-CoV-2 RNA not detected. Final results were converted into a .csv file with sample ID and its corresponding result and automatically uploaded into the DCL LIMS. During DCL operation, the Cq threshold defining the boundary between detected and indeterminate changed. For example, when there was a prevailing low population incidence of SARS-CoV-2, all positives were manually retested with a commercial three-plex (N-gene, Orf1a and S-gene targets) assay (TaqPath COVID-19 CE-IVD RT-PCR Kit, ThermoFisher). When there was a high prevalence, a Cq threshold of 32 was applied only above which was the TaqPath assay used. Results were evaluated manually and reported into the DCL LIMS as detected if two or more of the three TaqPath assays returned positive results, indeterminate if one of the assays was positive and negative if all three were negative.
Result reporting
Uploaded results encompassing an entire nominal roll triggered the LIMS to generate a report for that cohort. Results were reviewed by an on-call BMS or military consultant microbiologist. For indeterminate results, this clinical oversight allowed evaluation of reaction data and patient case history between the on-call military consultant and requesting MO to determine if an individual required a retest. Reports were emailed to requesting MOs or clinical staff at the PJHQ Co-ord Cell who were able to generate travel certificates. A summary of all daily positive cases was sent to the Defence Public Health Unit to initiate test and trace activities for military contacts. The patient details of these individuals were also uploaded to PHE (via DataMart) to facilitate test and trace activities for civilian contacts.
Supporting functions
Leadership team
A project technical authority and lead technical reviewer initiated or oversaw all developments. Ops managers liaised with the PJHQ COVID-19 Co-ord Cell, set up rotas and ensured these were resourced. A project manager oversaw all financial and contractual arrangements. Clinical governance was provided by Defence Consultant Advisor Pathology (DCA Path) and other military consultant microbiologists. To ensure continued staff engagement, the leadership team maintained communications with other DCL teams and staff through regular meetings and email updates.
Facilities and waste
Eight research CL2 laboratories were repurposed within the Dstl microbiological containment building. DCL operations were greatly helped by the building managers and staff of this facility. Three shipping container laboratories formed an admin hub and sample reception. Autoclaves were certified against the clinical waste streams. Autoclaved waste was incinerated.
Quality
Processes were validated against reference standards, external quality assessment (EQA) panels and previously tested samples from Frimley NHS Trust. In October 2020, following a review of validation reports and an assessment of protocols and processes, the United Kingdom Accreditation Service awarded extension to scope (under ISO/IEC 17025:2017, general requirements for the competence of testing and calibration laboratories framework) for the provision of a SARS-CoV-2 screening service in nasal swab samples. Accreditation was maintained by participation in laboratory EQA schemes, biweekly quality meetings, and a near-miss and non-conformancy reporting system.
Safety and security
A risk assessment was implemented with operational oversight by the Dstl Biological Safety Authority. Safety was continuously reviewed through an internal Dstl yellow card system. An anticontamination protocol (ie, prohibiting staff from PCR entering any other DCL lab and vice versa) was implemented. Personal security and operational security considerations were overseen by a security lead.
Training
The Dstl Chemical Biological Radiological (CBR) Specialist Training and Advisory team developed online and practical training packages for each role. Staff were required to have demonstrated competence in their roles before being signed off, and an online system was developed to ensure staff could demonstrate continued competency.
Technical support
Staff ensured labs were stocked with consumables and that stock levels were maintained. Sampling consumables were also supplied by the DCL to some units. Consumables were stored in shipping containers.
Laboratory results
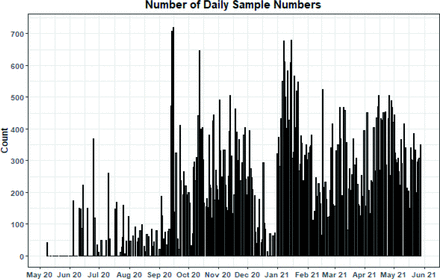
The DCL was operational on 289 days from May 2020 to May 2021 with a sustained 7-day-a-week capability operating from September 2021. More than 72 000 samples were processed (Table 1), with daily sample numbers flexing above the agreed 500 sample per day capacity on occasion (Figure 2). Of samples that cleared sample preparation, only 64 (0.09%) were determined as inhibited (void) after a retest. Six hundred military SARS-CoV-2 RNA-positive results were reported, with an E-gene PCR median Cq value of 30.44 (Table 2). Increased numbers of positives from September 2020 (Figure 3) apparently tracked the second wave of COVID-19 infections in the UK,12 with lowest median Cq values at the height of the second wave in January 2021 (Figure 4), potentially indicating the spread of the infectious alpha SARS-CoV-2 variant (which circulated in the UK from November 2020) within military populations. Throughout operations, positive samples with Cq values in the teens were observed (with occasional samples <13), indicating probable high viral loads in some individuals despite most testing being from ostensibly asymptomatic cohorts.
Summary of Defence Clinical Lab sample numbers and results
Summary of E-gene PCR Cq values from SARS-CoV-2 RNA detected samples
Daily sample numbers processed by DCL. DCL, Defence Clinical Lab.
Daily SARS-CoV-2 RNA detected sample numbers processed by DCL. DCL, Defence Clinical Lab.
Boxplot of Cq results from monthly SARS-CoV-2 RNA detected results. The monthly results from months with 10 or more positives only are shown. The box shows the IQR (25%–75%) along with the median; the whiskers show the range of the data excluding any outliers which are highlighted in red. Additionally, the mean has been added to the plot as a blue asterisk to highlight that the distributions each month were not normally distributed—this will mainly be due to the upper threshold. Cq, quantitation cycle.
Observations on laboratory operations
A majority of nominal rolls were submitted with at least a minor discrepancy or formatting issue, or as multiple versions of the same roll. This delayed analysis while admin staff reformatted rolls or verified information. Rolls were also submitted with non-clinical personnel listed as the requesting MO, causing checks on the clinical status of individuals to prevent transgressions of Caldicott principles. Patient information on swab tubes (usually hand written) also often differed from that provided on rolls delaying processing as these deviations were addressed.
Occurrences of incorrect packaging were also encountered. Global shortages of UN3373 compliant packaging led to some swab tubes being shipped in ziplock sandwich bags which were sometimes difficult to seal properly or were prone to open during transit. While such samples were still contained within primary packaging (ie, cardboard boxes), the reporting of such instances through the Dstl safety system, including staff unease in opening sample boxes on an open bench in the original sample reception protocol, led to the implementation of the prescreening sample reception procedure, within an MSC, where packaging was opened and verified as safe. Issues with packaging were recorded in an internal spreadsheet in order to provide feedback to units who were prone to sending such samples. A small number of cases were reported through the MOD Automated Significant Event Report system and samples were destroyed.
A potential point of failure was the use of a single target (E-gene) as the initial PCR. Although the three-plex TaqPath assay was used to confirm results, a danger persisted that variants evolving away from the E-gene target could have led to false-negative results. Although open-source material, PHE reports and operational data were analysed to identify any instances (with no issues observed), the assay was not changed due to the burdens of laboratory operation and uncertainty as to when the DCL would cease operation. However, an alternative duplex target commercial assay (VIASURE SARS-CoV-2 Real Time PCR Detection Kit, CerTest BioTec) was evaluated and identified as a replacement assay if it was required.
Considerations for a future pandemic response
It is difficult to extrapolate all lessons learnt to a future pandemic as the nature of different pathogens or advances in diagnostic technology would define a different response. However, some pointers as to how a similar military testing capability might be improved include the following.
Pan-MoD LIMS
UK academic laboratories which formed part of the UK Pillar 2 response tested sample tubes barcoded at the point of sampling, facilitating batching of samples onto liquid handling robots early in the laboratory process and rapid input of data into a LIMS.13 As discussed, the DCL expended much effort in verifying information on nominal rolls and handwritten on tubes at times, pushing the boundaries between a clinical requestor and clinical lab. A pan-MoD system where barcoded sample tubes were scanned and booked in at sampling would have alleviated many of these issues and facilitated greater automation of laboratory processes, reducing resource requirements while also increasing sample capacity.
Standardised or novel sampling consumables
An industry standard swab tube size and standardised packaging could have also facilitated laboratory automation. Swab transport media verified to inactivate SARS-CoV-2 but retain the RNA signature during transport from the sampling site to the laboratory could also have lowered sample inactivation requirements, reducing operative burdens and again facilitating increased throughput.
Conclusions
Through a sustained effort, the collaboration between Dstl scientific expertise and Defence Pathology clinical expertise provided the UK MoD with an accredited high-throughput SARS-CoV-2 diagnostic capability at the height of the COVID-19 pandemic. This was developed at pace and maintained for over 12 months with over 150 Dstl staff and military personnel working on the capability at some point. In testing over 72 000 samples—all associated with training, selection, operational deployments and overseas travel—the DCL significantly contributed to the maintenance of UK military capability and also reduced the risk of onward SARS-CoV-2 transmission within the defence community.
Although Dstl does not normally offer a reporting clinical diagnostic capability for infectious disease by collaborating with Defence Pathology, the DCL was able to offer a bespoke capability to the UK military with direct lines of communication to relevant stakeholders. The benefits of this included testing samples as unit or platform batches, retesting inhibited samples, providing agreed response timelines, requestor reachback to military consultant microbiologists and generally going the ‘extra mile’ with each sample. All these benefits may not have been available from other laboratories. Finally, the links between Dstl and Defence Pathology have been strengthened, benefitting future research and operational responses.
Ethics statements
Patient consent for publication
Acknowledgments
Previous projects in the Ministry of Defence Chief Scientific Advisor-funded Upstream Prevent, Protecting our People and CBR Services programmes greatly facilitated the rapid technical development of the capability. The authors thank colleagues from PHE Porton Down and representatives of Qiagen and ThermoFisher for help and assistance during the development phase. The authors also thank all Defence Science and Technology Laboratory staff and military personnel who worked for the Defence Clinical Lab during the development and operational phases.
Footnotes
Contributors SAW, SRA, SB, HB, EB, NC, KC, MC, JC, NAC, LC, VC, EE, CE, AF, CH, KH, GHarr, GHart, CH, BJ, HJ, EK, CL, LM, CM, JM, SN, MN, CO, AP, LP, RP, PR, VR, DR, FS, SS, HS, KStee, KStep, IT, JT, DU, NW, DW, ZW, CR and EJH significantly contributed to the initial development and/or operational maintenance of the capability. KC, SS, CH, MN and EJH provided clinical governance. VLC provided the statistical analysis. SAW wrote the paper with contributions from co-authors. All authors reviewed the manuscript for important intellectual content and approved the final version. SAW was the guarantor.
Funding The development and operation of the DCL was funded by the UK Department of Health and Social Care.
Competing interests None declared.
Provenance and peer review Not commissioned; externally peer reviewed.